
Volume 26, Number 1—January 2020
Research
Tick-Borne Encephalitis Virus, United Kingdom
On This Page
Downloads
Altmetric
Maya Holding , Stuart D. Dowall, Jolyon M. Medlock, Daniel P. Carter, Steven T. Pullan, James Lewis, Richard Vipond, Mara S. Rocchi, Matthew Baylis, and Roger Hewson
, Stuart D. Dowall, Jolyon M. Medlock, Daniel P. Carter, Steven T. Pullan, James Lewis, Richard Vipond, Mara S. Rocchi, Matthew Baylis, and Roger Hewson
Abstract
During February 2018–January 2019, we conducted large-scale surveillance for the presence and prevalence of tick-borne encephalitis virus (TBEV) and louping ill virus (LIV) in sentinel animals and ticks in the United Kingdom. Serum was collected from 1,309 deer culled across England and Scotland. Overall, 4% of samples were ELISA-positive for the TBEV serocomplex. A focus in the Thetford Forest area had the highest proportion (47.7%) of seropositive samples. Ticks collected from culled deer within seropositive regions were tested for viral RNA; 5 of 2,041 ticks tested positive by LIV/TBEV real-time reverse transcription PCR, all from within the Thetford Forest area. From 1 tick, we identified a full-length genomic sequence of TBEV. Thus, using deer as sentinels revealed a potential TBEV focus in the United Kingdom. This detection of TBEV genomic sequence in UK ticks has important public health implications, especially for undiagnosed encephalitis.
The only tickborne flavivirus in the United Kingdom documented to cause disease in vertebrates is louping ill virus (LIV), a vivirus transmitted by the deer/sheep tick, Ixodes ricinus (1). This tick species is the most abundant and widely distributed tick species in the United Kingdom and a known vector of Lyme borreliosis. LIV is most commonly detected in sheep, cattle, and red grouse and has been reported in Scotland, Wales, and England (primarily Cumbria, Devon, and North Yorkshire) (1). Humans are incidental hosts for LIV, and infection has been reported infrequently; ≈45 clinical cases have been linked to encephalitis during the past 85 years (1,2). However, the short window of acute infection leads to uncertainty about whether suspected cases resulted from LIV infection or some other cause, although serologic analysis to analyze recent exposure through induction of IgM-specific responses, in combination with clinical symptoms, could inform a presumptive diagnosis. Human cases are mostly linked to occupational exposure, particularly in abattoir or farm workers and occasionally in laboratory staff (2). Although the UK Animal and Plant Health Agency holds a database of confirmed diagnoses of LIV in livestock (3,4), the distribution and regional prevalence of LIV has not been fully defined. Records of distribution and regional prevalence are based on voluntary submissions by farmers and veterinarians from symptomatic livestock (1), from which private submissions are not integrated. Serologic analysis has been complicated; some animals received vaccination before its withdrawal.
Tick-borne encephalitis virus (TBEV) is a closely related flavivirus that, although known to be less virulent than LIV for sheep (5), causes a neurologic disease (tick-borne encephalitis [TBE]) after transmission to humans by infected ticks, producing clinical disease in an estimated one third of TBEV infections (6). TBE typically has a biphasic course starting with a prodromal phase with influenza-like symptoms, followed by a symptom-free interval before neurologic disease occurs; neurologic disease ranges from mild meningitis to severe encephalitis with or without myelitis and spinal paralysis (7). Three classic subtypes of TBEV are recognized: European (TBEV-Eu), Siberian, and Far Eastern. Two additional TBEV subtypes have recently been proposed: Baikalian subtype and the Himalayan subtype (8). TBEV-Eu is the prevailing subtype in Western Europe where it is primarily transmitted by I. ricinus ticks and is maintained within forest and meadow biotypes in endemic foci. In the United Kingdom, TBE is considered an imported disease; opportunities for the virus to become established principally are limited because the UK climate was not thought to support the specific conditions required for enzoonotic cycles to be established for TBEV to become endemic (9). However, changes in climate have affected the emergence, distribution, and abundance of I. ricinus in the United Kingdom (10); thus, the risk for tickborne disease has increased (11). A recent study provided evidence that co-infestation of tick larvae and nymphs occurs in small mammals in UK woodland (12). The increasing range of TBEV in Western Europe was underscored recently when the Netherlands reported its first human case in 2016 (13). Moreover, retrospective serologic screening of deer serum samples and molecular analysis of questing ticks found evidence of TBEV circulation in the Netherlands as far back as 2010 and 2015 (13,14). Given the increasing possibility that TBEV could be circulating in the United Kingdom, Public Health England developed a surveillance program focusing on wild animals and ticks.
In TBEV-endemic areas in continental Europe, the prevalence of TBEV in questing ticks is low, rarely exceeding 1% even in regions where the incidence of human infections is high (15). Therefore, instead of screening ticks directly, we used sentinel animals first to identify serologic evidence of TBEV to highlight sites for focused tick testing by specific TBEV detection using real-time reverse transcription PCR (rRT-PCR). Deer are proven as reliable sentinels for identifying areas where TBEV is present (13,15) because they have a limited home range, are available in large numbers, and are broadly dispersed within the surveillance areas. They also show long-lasting antibody responses after natural exposure to flaviviruses (15,16).
For our study, collectors retrieved blood samples from deer culled in England and Scotland during February 2018–January 2019; when available, they also collected tick samples. We tested the blood samples for TBEV or LIV antibodies and the ticks for the presence of viral RNA by rRT-PCR.
Sample Collection
We recruited persons involved in routine management of deer from across the United Kingdom to collect serum and tick samples from any species of deer. This program was promoted through organizations involved in deer management. These deerstalkers submitted 1,323 serum samples (and tick samples where present) from deer culled in England and Scotland during February 2018–January 2019. The University of Liverpool Ethics Committee (ref: VREC596) granted ethics approval for this study on February 1, 2018.
Blood samples were collected in serum-separation vacutainers from the chest cavity during gralloching, and blood-fed ticks were collected from any location on the deer carcass. Samples were centrifuged at 1,500 relative centrifugal force for 10 min and aliquoted. Serum and tick samples were stored at −80°C until further processing.
ELISA Testing
We tested serum samples for antibodies to TBEV using the commercial Immunozym FSME IgG All Species ELISA (Progen, ) according to the manufacturer’s instructions. We read plates at an optical density ratio of 450 nm. We considered samples with a reading of >127 Vienna units/mL to be seropositive.
Hemagglutination Inhibition Testing
Tick Identification and RNA Extraction
We morphologically identified all ticks collected from culled deer within a 15-km radius of any TBEV ELISA–seropositive deer (19) to life stage and species level. We individually homogenized the ticks in 300 μL RLT buffer (QIAGEN, ) in MK28-R Precellys homogenizing tubes using a Precellys 24 homogenizer (Bertin, ) at 5,500 rpm for 5 sec, followed by a 30-sec break; we repeated this process 4 times. We then added 300 μL of isopropanol and passed the tick homogenate through a QIAshredder (QIAGEN). We extracted total RNA using the BioSprint 96 One-For-All Vet Kit (QIAGEN) and eluted it into 100 μL AVE buffer according to the manufacturer’s instructions.
rRT-PCR
We tested individual tick samples for LIV/TBEV RNA using a sensitive LIV/TBEV assay (20). We amplified RNA in 20 μL rRT-PCR mix containing 0.8 μL Invitrogen () Superscript III/Platinum Taq Mix, 10 μL Invitrogen 2X reaction mix, 1.6 μL 50 mmol/L MgSO4,, 1 μL of 1 μmol/L forward primer, 1 μL of 18 μmol/L reverse primer, 0.2 μL of 25 μmol/L probe, 5 μL template, and 0.4 μL molecular-grade water.
We also tested all RNA-positive samples using a secondary assay designed to detect only LIV (21). We amplified RNA in 20 μL rRT-PCR mix containing 0.8 μL Invitrogen Superscript III/Platinum Taq Mix, 10 μL Invitrogen 2X reaction mix, 0.8 μL of 10 μmol/L forward primer, 1.8 μL of 10 μmol/L reverse primer, 1.0 μL of 5 μmol/L probe, 5 μL template, and 0.6 μL molecular-grade water.
Sequencing and Phylogenetic Analysis
We prepared the tick sample that showed a high level of TBEV RNA for metagenomic RNA sequencing (22) and assembled the sequencing data using SPAdes version 3.1.1 (23). We inferred the evolutionary history by using the maximum-likelihood method based on the Tamura 3-parameter model (24). We used the tree with the highest log likelihood. We automatically obtained initial trees for the heuristic search by applying neighbor-joining and BioNJ (25) algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value. The analysis involved 10 full-length genomic TBEV nucleotide sequences and was performed using Molecular Evolutionary Genetics Analysis version 7.0 software (26).
Deerstalkers submitted a total of 1,323 serum samples, of which 14 samples were excluded from analysis because of insufficient location or deer species information. Serum samples were obtained from 5 deer species and a hybrid of 2 species; 61% of samples submitted were from male deer. The most frequently sampled species were roe deer (Capreolus capreolus) (51%), followed by fallow deer (Dama dama) (19%). Samples were submitted from across Scotland and England, but distribution and density of samples varied by county (Figure 1, panel C). A limited number of samples were submitted from across the Midlands and parts of Northern England; no samples were submitted from Wales.
Of serum samples from across the United Kingdom, 4% were positive by ELISA, and 5% by HAI. Cohen’s κ indicated substantial agreement (0.61) between the methods, indicating ELISA results agreed closely with HAI test results (Table 1). ELISA yielded positive results in all deer species for which it was used. These were 27/663 roe, 10/246 fallow, 9/242 red deer (Cervus elaphus), 6/108 muntjac (Muntiacus reevesi), 1/48 sika (Cervus nippon), and 0/2 red/sika hybrids. HAI determined the following positives: 28/662 roe, 15/245 fallow, 18/242 red, 7/106 muntjac, 0/45 sika, and 1/2 red/sika hybrid.
ELISA- and HAI-positive samples were geographically distributed to specific areas (Figure 1, panels A, B); seroprevalence was high in southwestern Norfolk and northwestern Suffolk (Thetford Forest) in the east of England. Norfolk had the highest seroprevalence detected by ELISA (51.4%), followed by Hampshire (14.3%), Suffolk (10.7%), and Scottish Highlands (8.6%) (Table 2).
Of all ticks submitted from deer carcasses, 2,041 collected from 339 deer were from within 15 km of an ELISA-positive result. All ticks were identified as I. ricinus; 1,450 were adult females, 585 adult males, and 6 nymphs. Tick availability for testing by area of seropositive foci varied (Figure 1, panel C); most ticks tested were collected from Argyll and Bute, and an average of 6 ticks were tested per deer. Five (4 adult males, 1 adult female) of the 2,041 ticks tested positive by the LIV/TBEV rRT-PCR (20) and were all within the Norfolk/Suffolk focus (Figure 1, panel C). No LIV RNA was detected in these 5 ticks when they were tested by rRT-PCR designed to detect only LIV (21). The 192 ticks tested from within the Norfolk/Suffolk focus resulted in a prevalence of 2.6% in this area.
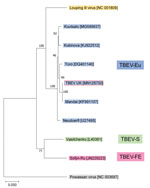
One tick (male) showed high levels of TBEV RNA (cycle threshold 15.4). Sequencing revealed a full-length TBEV genome designated TBEV-UK (GenBank accession no. MN128700). Phylogenetic analysis illustrates this as a TBEV-Eu subtype; it is most closely related to the Norwegian Mandal strain of TBEV isolated from ticks in 2009 (Figure 2), sharing a 99% sequence identity.
The detection of TBEV in the United Kingdom is important because TBEV can infect humans, causing febrile illness and neurologic complications including encephalitis. This evidence is contrary to earlier predictions based on climate change (9) that did not forecast a spread of TBEV to the United Kingdom. However, in addition to climate change, the spread of the tick vector, TBEV, and associated viruses into new regions can be influenced by a variety of other factors, such as transportation of animals and alterations in land management (27).
Serologic evidence suggests a high prevalence (47.7%) of exposure of deer to flaviviruses, such as TBEV and LIV, in the Norfolk/Suffolk (Thetford Forest) focal area. This seroprevalence is within the upper levels detected in TBEV risk areas of Europe, where seroprevalence studies in deer rarely exceed 50% (15,16,28,29). In addition, the detected prevalence of flavivirus RNA in ticks collected from deer of 2.6% within the Thetford Forest area falls within the range of findings from other studies in mainland Europe that tested blood-fed ticks (15). The deer were culled within a large forest habitat, which aligns closer with ecology required for TBEV, rather than LIV, maintenance (7). Based on these findings, and the evidence that all rRT-PCR–positive results were for TBEV and not LIV, we propose that TBEV is established and is being maintained through enzootic cycles within the Thetford Forest area, rather than resulting from multiple importation events, which is in line with findings in many endemic focal areas of TBEV (30). The hypothesis that TBEV infection might be maintained in Thetford Forest is supported by previous work in the United Kingdom that provided evidence of co-feeding between ticks from different life stages on small mammals in a southern English woodland, which is a crucial factor for the maintenance of TBEV (9,12). In addition, our positive serology data support the concept that the virus is circulating nonviremically in local wildlife and by cycling among the co-feeding nymphs, larvae, and adult ticks, through nonviremic transmission. Good evidence shows that TBEV is maintained in other parts of Europe through nonviremic transmission (31).
In other study areas, we detected serologic evidence of flavivirus exposure but not viral RNA in ticks. The close homology between LIV and TBEV presents challenges when serologic methods are used alone because the tests cannot distinguish between them. Thus, based on such data, confirming which virus is responsible for the seroreactivity in the areas where LIV has previously been reported is not possible. Previous reports of LIV prevalence are limited; just one study showed up to 15.3% of ticks positive for LIV (32). However, other researchers have not confirmed these data, and our results indicate a much lower prevalence. Nevertheless, we did not find any published clinical reports of LIV in Hampshire livestock despite our detected seroprevalence of 14.3% by ELISA (1,3,4). Although additional tick and small mammal ecology studies are needed to build on serologic data, evidence shows the maintenance of TBEV in the identified focal endemic area.
The genomic sequence of TBEV-UK shows close identity to a TBEV-Eu virus isolated in 2009 from questing ticks collected in Norway. This similarity suggests that TBEV-UK might have been brought to the United Kingdom on migratory birds, such as blackbirds (Turdus merula) and redwings (Turdus iliacus) (33), which are known to transport ticks over wide distances (34–37). The United Kingdom experiences a large influx of migratory birds each autumn from several TBEV-endemic countries in northern Europe, including Norway. During this migration, birds first arrive on the east coast of the United Kingdom, and it is feasible that TBEV-UK could have originated from a tick imported by an autumn migratory bird. We are collecting tick samples from migratory birds to assess the proportion of tick-infested birds arriving in the United Kingdom and testing these imported ticks for TBEV (among other potential pathogens). In addition, because of lower viral RNA levels, we are looking into primer-amplification sequencing approaches to further decipher the virus responsible for the rRT-PCR–positive samples detected from Thetford Forest.
For zoonotic infections, detection of a pathogen in the animal reservoir/host, vector, or both often precedes the emergence of human infection (38). Such was the case in the Netherlands, where deer serum samples, collected 6 years before the first cases in humans (13), demonstrated serologic evidence of TBEV infection (14). Similarly, in Spain, Crimean-Congo hemorrhagic fever virus was first reported in ticks in 2010 (39), before autochthonous infections in humans was identified in 2017 (40). Within the focal TBEV-endemic areas we identified in this study, seroepidemiologic studies should be undertaken, particularly in risk groups that include patients presenting to general practitioners and hospitals with central nervous system symptoms.
Although UK-TBEV has not been linked to human disease, it nevertheless shows close homology to pathogenic isolates of TBEV and should be considered to be a potential public health risk. Thus, clinicians in the United Kingdom should consider the European Union case definition of TBE (41) and include TBE in the differential diagnosis of patients with symptoms of meningoencephalitis, especially if they have been exposed to a tick bite, even if they have not traveled recently to a known TBE-endemic country. The European Union case definition specifies clinical criteria as any person presenting with inflammation of the central nervous system. In addition to meeting clinical criteria, laboratory case confirmation requires >1 of the following 5 criteria to be satisfied: 1) detection of TBEV nucleic acid, 2) viral isolation from clinical specimens, 3) TBEV-specific IgM and IgG in blood, 4) TBEV-specific IgM in cerebrospinal fluid, and 5) seroconversion or 4-fold increase of TBEV-specific antibodies in paired serum samples (41).
Although no autochthonous cases of clinical human disease have been diagnosed in the United Kingdom, up to 60% of encephalitis cases reach no diagnosis (42). Therefore, our results indicate that TBEV should be considered as a potential cause in encephalitis patients, and the wide distribution of the natural vector in the United Kingdom indicates a need for close monitoring and a potential for geographic spread and expanding risk areas.
Ms. Holding is a PhD candidate at Public Health England. Her primary research interests are the ecology and epidemiology of vectorborne pathogens.
Acknowledgments
We are grateful to the Forestry Commission, British Deer Society, Deer Initiative, and Barry Dowsett for their support and promotion of this study. We thank all of the participating deerstalkers for their support in collecting samples for this study; Dylan Turnbull and Ellie Laming for performing HAI testing; Gillian Slack, John Chamberlain, Emma Kennedy, and Victoria Graham for their help processing samples; Penny Roberts for producing study packs; Hein Sprong for his advice in setting up the study; and Nick Johnson for providing control serum for assay testing.
This research was funded by the Virology & Pathogenesis Group’s Arbovirus Programme, Public Health England, and by the National Institute for Health Research Health Protection Research Unit in Emerging and Zoonotic Infections at the University of Liverpool in partnership with Public Health England, in collaboration with Liverpool School of Tropical Medicine. M.H. is funded by the National Institute of Health Research Health Protection Research Unit in Emerging and Zoonotic Infections. M.S.R. is funded by the Scottish Government.
References
- Jeffries CL, Mansfield KL, Phipps LP, Wakeley PR, Mearns R, Schock A, et al. Louping ill virus: an endemic tick-borne disease of Great Britain. J Gen Virol. 2014;95:1005–14.
- Davidson MM, Williams H, Macleod JAJ. Louping ill in man: a forgotten disease. J Infect. 1991;23:241–9.
- Animal & Plant Health Agency; Scotland’s Rural College. GB sheep disease surveillance dashboard [cited 2019 May 20].
- Animal & Plant Health Agency; Scotland’s Rural College. GB cattle disease surveillance dashboard [cited 2019 May 20].
- Gritsun TS, Nuttall PA, Gould EA. Tick-borne flaviviruses. Adv Virus Res. 2003;61:317–71.
- European Centre for Disease Prevention and Control. Epidemiological situation of tick-borne encephalitis in the European Union and European Free Trade Association countries. ECDC Technical Report. 2012 [cited 2019 Jun 6].
- Lindquist L, Vapalahti O. Tick-borne encephalitis. Lancet. 2008;371:1861–71.
- Dai X, Shang G, Lu S, Yang J, Xu J. A new subtype of eastern tick-borne encephalitis virus discovered in Qinghai-Tibet Plateau, China. Emerg Microbes Infect. 2018;7:74.
- Randolph SE, Rogers DJ. Fragile transmission cycles of tick-borne encephalitis virus may be disrupted by predicted climate change. Proc Biol Sci. 2000;267:1741–4.
- Medlock JM, Hansford KM, Vaux AGC, Cull B, Gillingham E, Leach S. Assessment of the public health threats posed by vector-borne disease in the United Kingdom (UK). Int J Environ Res Public Health. 2018;15:2145.
- Medlock JM, Leach SA. Effect of climate change on vector-borne disease risk in the UK. Lancet Infect Dis. 2015;15:721–30.
- Cull B, Vaux AGC, Ottowell LJ, Gillingham EL, Medlock JM. Tick infestation of small mammals in an English woodland. J Vector Ecol. 2017;42:74–83.
- Jahfari S, de Vries A, Rijks JM, Van Gucht S, Vennema H, Sprong H, et al. Tick-borne encephalitis virus in ticks and roe deer, the Netherlands. Emerg Infect Dis. 2017;23:1028–30.
- de Graaf JA, Reimerink JH, Voorn GP, Bij de Vaate EA, de Vries A, Rockx B, et al. First human case of tick-borne encephalitis virus infection acquired in the Netherlands, July 2016. Euro Surveill. 2016;21:30318.
- Imhoff M, Hagedorn P, Schulze Y, Hellenbrand W, Pfeffer M, Niedrig M. Review: Sentinels of tick-borne encephalitis risk. Ticks Tick Borne Dis. 2015;6:592–600.
- Kiffner C, Vor T, Hagedorn P, Niedrig M, Rühe F. Determinants of tick-borne encephalitis virus antibody presence in roe deer (Capreolus capreolus) sera. Med Vet Entomol. 2012;26:18–25.
- Ytrehus B, Vainio K, Dudman SG, Gilray J, Willoughby K. Tick-borne encephalitis virus and louping-ill virus may co-circulate in Southern Norway. Vector Borne Zoonotic Dis. 2013;13:762–8.
- Clarke DH, Casals J. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am J Trop Med Hyg. 1958;7:561–73.
- Hillyard PD. Ticks of north-west Europe. Shrewsbury (UK): Field Studies Council; 1996.
- Schwaiger M, Cassinotti P. Development of a quantitative real-time RT-PCR assay with internal control for the laboratory detection of tick borne encephalitis virus (TBEV) RNA. J Clin Virol. 2003;27:136–45.
- Marriott L, Willoughby K, Chianini F, Dagleish MP, Scholes S, Robinson AC, et al. Detection of Louping ill virus in clinical specimens from mammals and birds using TaqMan RT-PCR. J Virol Methods. 2006;137:21–8.
- Kafetzopoulou LE, Efthymiadis K, Lewandowski K, Crook A, Carter D, Osborne J, et al. Assessment of metagenomic Nanopore and Illumina sequencing for recovering whole genome sequences of chikungunya and dengue viruses directly from clinical samples. Euro Surveill. 2018;23:
1800228 . - Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, et al. Assembling genomes and mini-metagenomes from highly chimeric reads. In: Deng M, Jiang R, Sun F, Zhang X, editors. Research in computational molecular biology. RECOMB 2013. Lecture notes in computer science, vol 7821. Berlin: Springer; 2013. p. 158–70.
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. 1992;9:678–87.
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–95.
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
- Gould EA, Higgs S, Buckley A, Gritsun TS. Potential arbovirus emergence and implications for the United Kingdom. Emerg Infect Dis. 2006;12:549–55.
- Duscher GG, Wetscher M, Baumgartner R, Walder G. Roe deer sera used for TBE surveillance in Austria. Ticks Tick Borne Dis. 2015;6:489–93.
- Tavernier P, Sys SU, De Clercq K, De Leeuw I, Caij AB, De Baere M, et al. Serologic screening for 13 infectious agents in roe deer (Capreolus capreolus) in Flanders. Infect Ecol Epidemiol. 2015;5:29862.
- Burton A. Tick-borne encephalitis emerges in Norway. Lancet Infect Dis. 2004;4:189.
- Labuda M, Jones LD, Williams T, Danielova V, Nuttall PA. Efficient transmission of tick-borne encephalitis virus between cofeeding ticks. J Med Entomol. 1993;30:295–9.
- Watts EJ, Palmer SCF, Bowman AS, Irvine RJ, Smith A, Travis JMJ. The effect of host movement on viral transmission dynamics in a vector-borne disease system. Parasitology. 2009;136:1221–34.
- Sparagano O, George D, Giangaspero A, Špitalská E. Arthropods and associated arthropod-borne diseases transmitted by migrating birds. The case of ticks and tick-borne pathogens. Vet Parasitol. 2015;213:61–6.
- Waldenström J, Lundkvist A, Falk KI, Garpmo U, Bergström S, Lindegren G, et al. Migrating birds and tickborne encephalitis virus. Emerg Infect Dis. 2007;13:1215–8.
- Geller J, Nazarova L, Katargina O, Leivits A, Järvekülg L, Golovljova I. Tick-borne pathogens in ticks feeding on migratory passerines in Western part of Estonia. Vector Borne Zoonotic Dis. 2013;13:443–8.
- Hasle G. Transport of ixodid ticks and tick-borne pathogens by migratory birds. Front Cell Infect Microbiol. 2013;3:48.
- Klaus C, Gethmann J, Hoffmann B, Ziegler U, Heller M, Beer M. Tick infestation in birds and prevalence of pathogens in ticks collected from different places in Germany. Parasitol Res. 2016;115:2729–40.
- Randolph SE, Sumilo D. Tick-borne encephalitis in Europe: dynamics of changing risk. In: Takken W, Knols BGJ, editors Emerging pests and vector-borne diseases in Europe. Wageningen (the Netherlands): Wageningen Academic Publishers; 2007. p. 187–206.
- Estrada-Peña A, Palomar AM, Santibáñez P, Sánchez N, Habela MA, Portillo A, et al. Crimean-Congo hemorrhagic fever virus in ticks, Southwestern Europe, 2010. Emerg Infect Dis. 2012;18:179–80.
- Negredo A, de la Calle-Prieto F, Palencia-Herrejón E, Mora-Rillo M, Astray-Mochales J, Sánchez-Seco MP, et al.; Crimean Congo Hemorrhagic Fever@Madrid Working Group. Autochthonous Crimean-Congo Hemorrhagic Fever in Spain. N Engl J Med. 2017;377:154–61.
- European Commission. Commission implementing decision of 8 August 2012 amending Decision 2002/253/EC laying down case definitions for reporting communicable diseases to the Community network under Decision No 2119/98/EC of the European Parliament and of the Council. Official Journal of the European Union. 2012;55(L 262/1):1–57.
- Kennedy PGE, Quan P-L, Lipkin WI. Viral encephalitis of unknown cause: current perspective and recent advances. Viruses. 2017;9:138.
Figures
Tables
Cite This ArticleOriginal Publication Date: 10/29/2019

No comments:
Post a Comment