
Volume 25, Number 5—May 2019
Dispatch
Rickettsia japonica and Novel Rickettsia Species in Ticks, China
On This Page
Xiang-Rong Qin, Hui-Ju Han, Fu-Jun Han, Fu-Ming Zhao, Zhen-Tang Zhang, Zai-Feng Xue, Dong-Qiang Ma, Rui Qi, Min Zhao, Li-Jun Wang, Li Zhao, Hao Yu, Jian-Wei Liu , and Xue-Jie Yu
, and Xue-Jie Yu
Abstract
PCR amplification indicated the minimum infection rate of Rickettsia spp. was 0.66% in Haemaphysalis longicornis ticks collected from Shandong Province, China. Phylogenetic analysis based on the rrs, gltA, ompA, and ompB genes indicated that the ticks carried R. japonica, CandidatusRickettsia longicornii, and a novel Rickettsia species related to R. canadensis.
Rickettsia species are gram-negative obligate intracellular bacteria that infect humans and a variety of vertebrates through the bite of arthropod vectors. Hard-body ticks are the primary vector of spotted fever group (SFG) rickettsiae; recently, several emerging and reemerging SFG rickettsiae were found to infect humans (1). Rickettsia japonica is the pathogenic agent of Japanese spotted fever that has been reported in Japan, South Korea, and Thailand since 1984 (2–4). Japanese spotted fever is a severe zoonosis and develops abruptly with headache, fever, shaking chills, skin eruptions, tick bite eschars, and malaise (2). R. canadensis was initially isolated from ticks in Canada; a serologic study indicated the presence of R. canadensis antibodies in febrile patients (5). The presence of Rickettsia species and their distributions in China are not very clear. In this study, we analyzed Rickettsia species in Haemaphysalis longicornis ticks collected from Shandong Province, China, and found R. japonica, Candidatus Rickettsia longicornii, and a novel Rickettsiaspecies closely related to R. canadensis in the ticks.
We collected questing ticks by flagging during April–July 2013–2015. We collected them in Jiaonan County (35°35′–36°8′ N and 119°30′–120°11′E), Shandong Province, China. Jiaonan County is located on the Pacific coast of China and has a maritime monsoon-type climate. We identified tick species individually by morphology and confirmed by PCR amplification and DNA sequencing of the 16S rRNA gene of 2 nymphs and 2 adult ticks of each species as described previously (6,7).
For detection of Rickettsia DNA, we pooled ticks according to their developmental stages, with each pool consisting of 20 nymphs or 10 adult ticks. We homogenized them with Tissue Lyser II (QIAGEN, http://www.qiagen.com). We extracted total nucleic acids from the tick suspension using the AllPrep DNA/RNA Mini Kit (QIAGEN).
Initially, in all the tick pools, we amplified nucleic acid preparations with rickettsial universal primers targeting rrs, gltA, and ompB (B1–B4). We further amplified Rickettsia clones in the tick pools closely related to R. japonica with primers of ompA, a SFG rickettsia unique gene. The clones positive with rrsand gltA gene primers but negative with ompB primers (B1–B4) we further amplified with primers Cand-1 to Cand-4, which were designed from the R. canadensis ompB gene because the Rickettsia clones from these tick pools were closely related to R. canadensis on the basis of the rrs and gltA gene sequences (Table). We used distilled water as a negative control in each run.
We performed electrophoresis on the PCR products in 1.2% agarose gels, stained them with ethidium bromide, and visualized them under UV light. DNA bands with the expected size were excised and extracted by Gel Extraction Kit (Omega Bio-tek, https://www.omegabiotek.com). We cloned the purified PCR products into pMD19-T vector (Takara, https://www.takara-bio.com) and engaged Sangon Biotech (Shanghai, China) (https://www.life-biotech.com) to conduct sequencing on both strands. We compared nucleotide sequences with BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and constructed a phylogenetic tree using the maximum-likelihood method with MEGA version 6.0 (https://www.megasoftware.net). We deposited the Rickettsia genes obtained in this study in GenBank under accession nos. MF496152–MF496168 (rrs), MF496169–MF496185 (gltA), MF496186–MF496199 (ompB), and MK102707–MK102720 (ompA).
We collected a total of 2,560 H. longicornis ticks, 2,080 nymphs and 480 adults. PCR amplification indicated that 14 tick pools were positive with rrs, gltA, and ompB (B1–B4) primers and further positively amplified by PCR with ompA primers. In addition, 3 clones were positive with rrs, gltA, and ompB (Cand-1 to Cand-4) primers. The minimum infection rate of Rickettsia in the ticks was 0.66% (17/2,560), assuming 1 tick was positive in each positive pool of ticks.
Sequence analysis indicated that 3 clones (J84, J85, and J217) detected from the tick pools were closely related to R. canadensis, showing sequence homology of 98.7%–99.1% for rrs, 97.8%–98.4% for gltA and 94.8%–95.1% for ompB. One clone (J244) was highly homologous to Candidatus Rickettsia longicornii, showing sequence homology of 99.2% for rrs, 100% for gltA, and 99.7% for ompA. The remaining 13 clones were homologous to each other and to R. japonica, showing sequence homology of 99. 2%–100% for rrs, 99.1%–100% for gltA, 99.3%–99.4% for ompB, and 97%–97.3% for ompA of a variety strains of R. japonica (Appendix Tables 1–4).
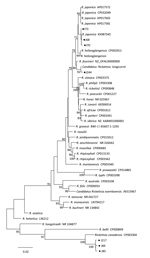
Figure. Phylogenetic tree of isolates from study of Rickettsia species in China (black dots) and comparison isolates. The tree was generated using the concatenated sequences of rrs, gltA, ompB, and ompA of...
Phylogenetic analysis based on the concatenated sequences of rrs, gltA, ompB, and ompA showed that Rickettsia clones (J84, J85, and J217) were clustered in the same clade with, but distinct from, R. canadensis; clone J244 was in the same clade as Candidatus Rickettsia longicornii; the remaining 13 clones were in the same clade as R. japonica. These results indicated that clones J84, J85, and J217 were a novel Rickettsia species; clone 244 was Candidatus Rickettsia longicornii; and other clones were R. japonica (Figure).
In this study, we demonstrated that H. longicornis ticks from China were infected with multiple Rickettsia species, including R. japonica, CandidatusRickettsia longicornii, and a novel Rickettsia species. We named the novel species Candidatus Rickettsia jiaonani after the sampling site. The exact classification of Candidatus Rickettsia jiaonani needs to be further studied by sequencing the whole genomes of the organisms.
R. japonica infection in humans has been reported recently in Anhui Province in central China (11), suggesting that R. japonica is widely distributed in China and its epidemiology needs to be further investigated. Candidatus Rickettsia longicornii was previously detected in H. longicornis ticks collected from South Korea (12). Candidatus Rickettsia jiaonani is closely related to R. canadensis, which was first isolated from H. leporispalustris ticks removed from rabbits in Ontario, Canada, in 1963 and then from a H. leporispalustris tick removed from a black-tailed jackrabbit in California in 1980 (13).
H. longicornis ticks are native to East Asia, including China, Korea, and Japan, and they were introduced into Oceania, including Australia, New Zealand, Fiji, and Hawaii, through cattle importation (6). Recently, this tick species was found in 8 states in the eastern United States (14). This study and previous studies demonstrated that H. longicornis ticks carry R. japonica, Candidatus Rickettsia longicornii, Candidatus Rickettsia jiaonani, Anaplasma phagocytophilum, Ehrlichia, and severe fever with thrombocytopenia syndrome virus (12,15). These pathogens need to be monitored in countries in East Asia in which the H. longicornis tick is native and in the countries that this tick species has invaded.
Ms. Qin is a PhD student in the School of Health Sciences of Wuhan University. Her research interest is infectious disease epidemiology.
Acknowledgment
This study was supported by a grant from the National Natural Science Funds of China (no. 31570167).
References
- Teng K, Jiang Z. Economic insect fauna of China Fasc 39 Acari: Ixodidae. Fauna Sinica Beijing: Science Press. 1991.
- Igolkina YP, Rar VA, Yakimenko VV, Malkova MG, Tancev AK, Tikunov AY, et al. Genetic variability of Rickettsia spp. in Ixodes persulcatus/Ixodes trianguliceps sympatric areas from Western Siberia, Russia: Identification of a new Candidatus Rickettsia species. Infect Genet Evol. 2015;34:88–93. DOIPubMed
- Roux V, Fournier PE, Raoult D. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J Clin Microbiol. 1996;34:2058–65.PubMed
Figure
Table
Cite This ArticleOriginal Publication Date: 3/26/2019
No comments:
Post a Comment